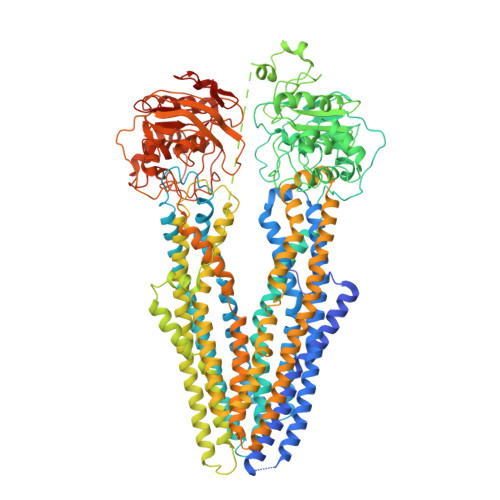
Structures of ABCB4 provide insight into phosphatidylcholine translocation.
Nosol, K., Bang-Sorensen, R., Irobalieva, R.N., Erramilli, S.K., Stieger, B., Kossiakoff, A.A., Locher, K.P.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34385322
- DOI: https://doi.org/10.1073/pnas.2106702118
- Primary Citation of Related Structures:
7NIU, 7NIV, 7NIW - PubMed Abstract:
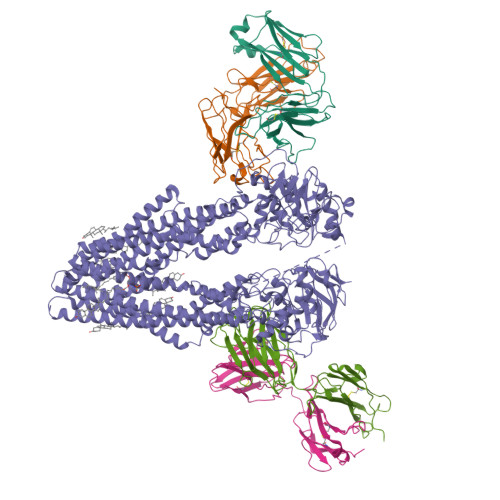
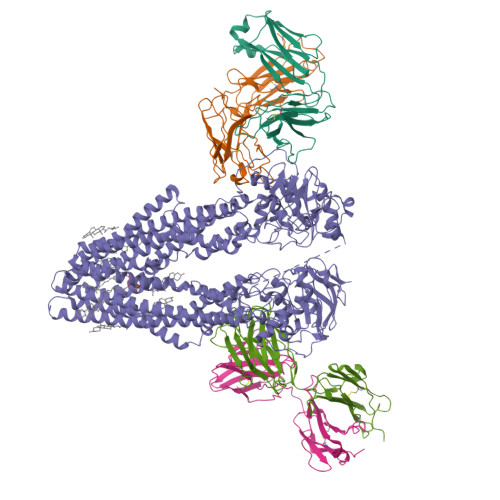
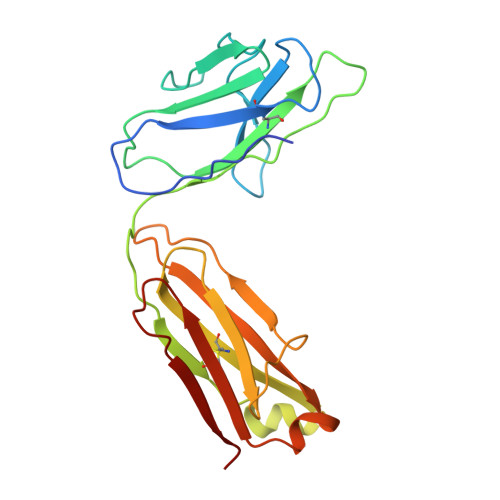
ABCB4 is expressed in hepatocytes and translocates phosphatidylcholine into bile canaliculi. The mechanism of specific lipid recruitment from the canalicular membrane, which is essential to mitigate the cytotoxicity of bile salts, is poorly understood. We present cryogenic electron microscopy structures of human ABCB4 in three distinct functional conformations. An apo-inward structure reveals how phospholipid can be recruited from the inner leaflet of the membrane without flipping its orientation. An occluded structure reveals a single phospholipid molecule in a central cavity. Its choline moiety is stabilized by cation-π interactions with an essential tryptophan residue, rationalizing the specificity of ABCB4 for phosphatidylcholine. In an inhibitor-bound structure, a posaconazole molecule blocks phospholipids from reaching the central cavity. Using a proteoliposome-based translocation assay with fluorescently labeled phosphatidylcholine analogs, we recapitulated the substrate specificity of ABCB4 in vitro and confirmed the role of the key tryptophan residue. Our results provide a structural basis for understanding an essential translocation step in the generation of bile and its sensitivity to azole drugs.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, ETH Zürich, 8093 Zürich, Switzerland.